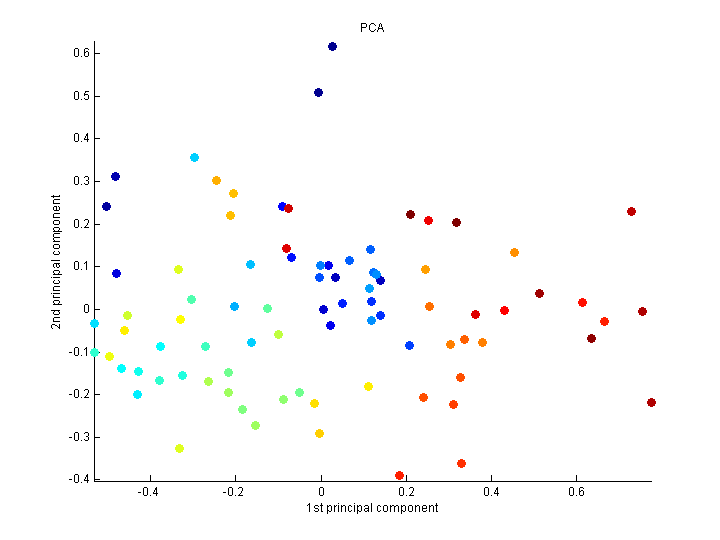
Each point is a row.



| 1 | 31 | 1869RR.cel | |||
| 2 | 29 | 1795R.cel | |||
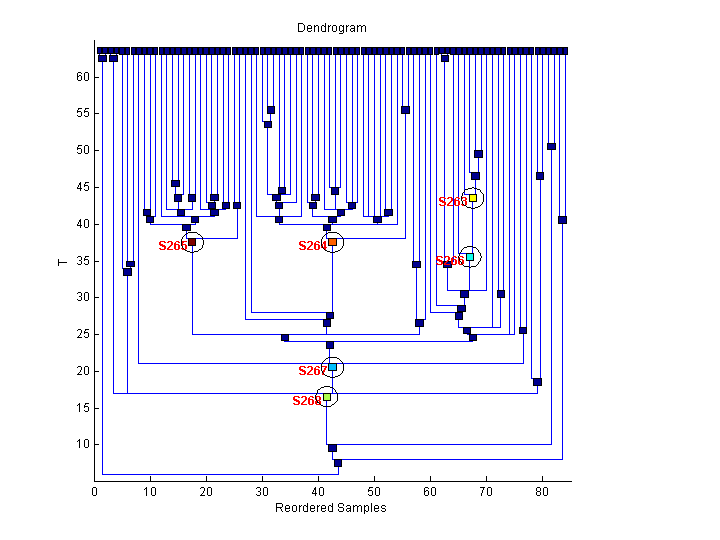
| 3 | 66 | S268 | 55.cel | ||
| 4 | 8 | S268 | 1297.cel | ||
| 5 | 25 | S268 | 161.cel | ||
| 6 | 33 | S268 | 211.cel | ||
| 7 | 68 | S268 | 563.cel | ||
| 8 | 35 | S267 S268 | 218.cel | ||
| 9 | 9 | S265 S267 S268 | 1308.cel | ||
| 10 | 13 | S265 S267 S268 | 1399.cel | ||
| 11 | 36 | S265 S267 S268 | 222.cel | ||
| 12 | 17 | S265 S267 S268 | 147.cel | ||
| 13 | 6 | S265 S267 S268 | 1284.cel | ||
| 14 | 26 | S265 S267 S268 | 1621.cel | ||
| 15 | 27 | S265 S267 S268 | 1622R.cel | ||
| 16 | 48 | S265 S267 S268 | 323.cel | ||
| 17 | 39 | S265 S267 S268 | 246.cel | ||
| 18 | 56 | S265 S267 S268 | 396.cel | ||
| 19 | 19 | S265 S267 S268 | 1497R.cel | ||
| 20 | 24 | S265 S267 S268 | 159.cel | ||
| 21 | 47 | S265 S267 S268 | 300.cel | ||
| 22 | 49 | S265 S267 S268 | 328.cel | ||
| 23 | 10 | S265 S267 S268 | 1317.cel | ||
| 24 | 57 | S265 S267 S268 | 428.cel | ||
| 25 | 53 | S265 S267 S268 | 356bis.cel | ||
| 26 | 82 | S265 S267 S268 | 97.cel | ||
| 27 | 1 | S267 S268 | 100R.cel | ||
| 28 | 11 | S267 S268 | 1357.cel | ||
| 29 | 20 | S264 S267 S268 | 15.cel | ||
| 30 | 18 | S264 S267 S268 | 148.cel | ||
| 31 | 4 | S264 S267 S268 | 1076.cel | ||
| 32 | 28 | S264 S267 S268 | 1780.cel | ||
| 33 | 22 | S264 S267 S268 | 155R.cel | ||
| 34 | 45 | S264 S267 S268 | 288.cel | ||
| 35 | 55 | S264 S267 S268 | 390.cel | ||
| 36 | 37 | S264 S267 S268 | 236.cel | ||
| 37 | 30 | S264 S267 S268 | 1854R.cel | ||
| 38 | 23 | S264 S267 S268 | 157.cel | ||
| 39 | 12 | S264 S267 S268 | 1360.cel | ||
| 40 | 70 | S264 S267 S268 | 57.cel | ||
| 41 | 32 | S264 S267 S268 | 190.cel | ||
| 42 | 38 | S264 S267 S268 | 242.cel | ||
| 43 | 40 | S264 S267 S268 | 254.cel | ||
| 44 | 43 | S264 S267 S268 | 283.cel | ||
| 45 | 16 | S264 S267 S268 | 1453.cel | ||
| 46 | 51 | S264 S267 S268 | 345.cel | ||
| 47 | 79 | S264 S267 S268 | 90.cel | ||
| 48 | 21 | S264 S267 S268 | 1553.cel | ||
| 49 | 50 | S264 S267 S268 | 344.cel | ||
| 50 | 63 | S264 S267 S268 | 528bis.cel | ||
| 51 | 75 | S264 S267 S268 | 70.cel | ||
| 52 | 15 | S264 S267 S268 | 1437.cel | ||
| 53 | 81 | S264 S267 S268 | 94bis.cel | ||
| 54 | 76 | S264 S267 S268 | 80.cel | ||
| 55 | 61 | S264 S267 S268 | 506.cel | ||
| 56 | 62 | S264 S267 S268 | 527.cel | ||
| 57 | 14 | S267 S268 | 1430.cel | ||
| 58 | 65 | S267 S268 | 547.cel | ||
| 59 | 84 | S267 S268 | ge205.cel | ||
| 60 | 2 | S267 S268 | 104.cel | ||
| 61 | 3 | S267 S268 | 106.cel | ||
| 62 | 83 | S267 S268 | ge197.cel | ||
| 63 | 69 | S267 S268 | 566.cel | ||
| 64 | 41 | S267 S268 | 269.cel | ||
| 65 | 54 | S266 S267 S268 | 360.cel | ||
| 66 | 42 | S263 S266 S267 S268 | 274.cel | ||
| 67 | 46 | S263 S266 S267 S268 | 30.cel | ||
| 68 | 58 | S263 S266 S267 S268 | 444.cel | ||
| 69 | 59 | S263 S266 S267 S268 | 458.cel | ||
| 70 | 60 | S267 S268 | 461.cel | ||
| 71 | 52 | S267 S268 | 349.cel | ||
| 72 | 5 | S267 S268 | 12.cel | ||
| 73 | 71 | S267 S268 | 574.cel | ||
| 74 | 73 | S267 S268 | 63.cel | ||
| 75 | 74 | S267 S268 | 65R.cel | ||
| 76 | 78 | S267 S268 | 84.cel | ||
| 77 | 80 | S267 S268 | 93.cel | ||
| 78 | 7 | S268 | 129.cel | ||
| 79 | 44 | S268 | 284.cel | ||
| 80 | 67 | S268 | 558.cel | ||
| 81 | 34 | 212R.cel | |||
| 82 | 72 | 57R.cel | |||
| 83 | 64 | 534.cel | |||
| 84 | 77 | 83.cel |
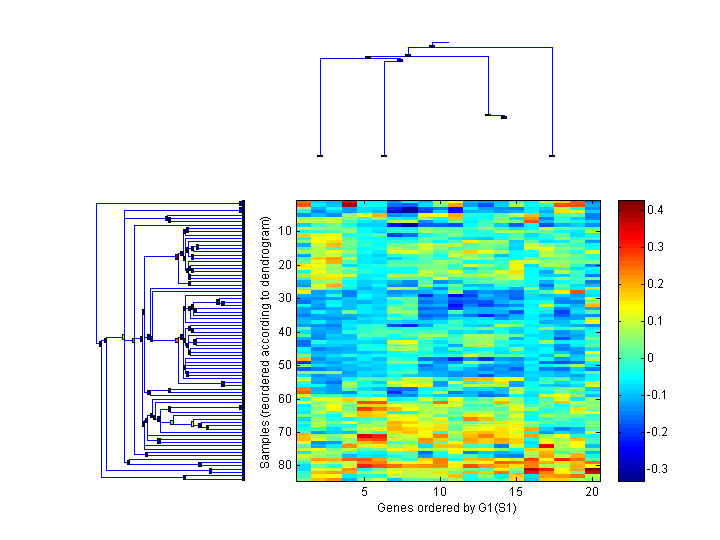
| 1 | 2 | 204121_at | GADD45G Chr:9q22.1-q22.2 | |
| 2 | 14 | 229400_at | HOXD10 Chr:2q31.1 | |
| 3 | 18 | 238847_at | --- --- | |
| 4 | 5 | 206858_s_at | HOXC6 Chr:12q13.3 | |
| 5 | 6 | 209905_at | HOXA9 Chr:7p15-p14 | |
| 6 | 9 | 214651_s_at | HOXA9 Chr:7p15-p14 | |
| 7 | 3 | 204362_at | SCAP2 Chr:7p21-p15 | |
| 8 | 10 | 225639_at | SCAP2 Chr:7p21-p15 | |
| 9 | 8 | 213844_at | HOXA5 Chr:7p15-p14 | |
| 10 | 7 | 213150_at | HOXA10 Chr:7p15-p14 | |
| 11 | 11 | 226582_at | --- Chr:12q13.13 | |
| 12 | 1 | 1557051_s_at | --- --- | |
| 13 | 13 | 228642_at | --- --- | |
| 14 | 16 | 235521_at | HOXA3 Chr:7p15-p14 | |
| 15 | 17 | 235753_at | --- --- | |
| 16 | 19 | 239153_at | --- Chr:12q13.13 | |
| 17 | 12 | 228564_at | LOC375295 Chr:2q31.2 | |
| 18 | 4 | 205522_at | HOXD4 Chr:2q31.1 | |
| 19 | 15 | 231906_at | HOXD8 Chr:2q31.1 | |
| 20 | 20 | 244521_at | --- --- |
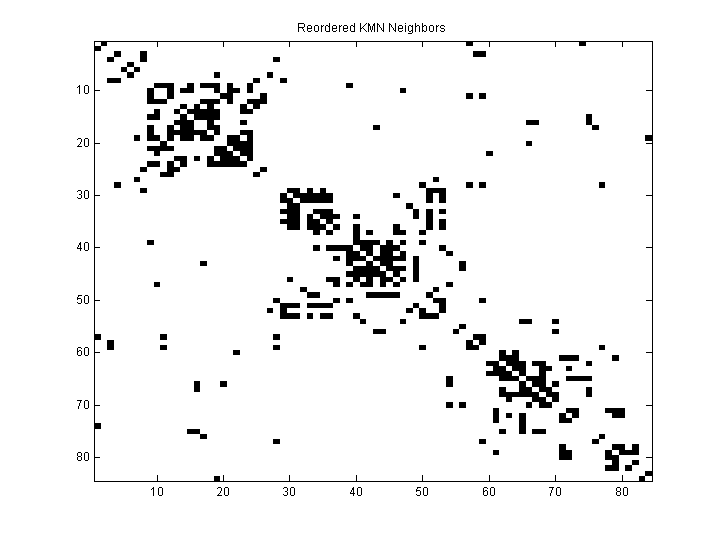
| K (nearest neighbors) | 10 |
| Min T | 0.000000 |
| Max T | 0.250000 |
| Delta T | 0.004000 |
| Cycles | 3000 |
| Growth | TRUE |
| Stable delta T | 4 |
| Ignore dropout size | 1 |